Research
During my Ph.D. study, I was working under the supervision of Dr. Enamul Huq, Plant Biology Graduate Program at UT
Current projects
My research interests focus on applying both molecular genetics and bioinformatics to understand the light signaling in Arabidopsis.

• Project 1: Applied negative binomial generalized linear models for different expression comparison and clustering analysis (k-means and hierarchical clustering) in R using gene expression data to reveal intricate network of developmental pathways regulated by key regulators in Arabidopsis.

• Project 2: Develop computational pipelines using Unix/R and statistical based methods using R to analyze coexpression data from 60 different transcriptome sequencing samples to understand transcriptional regulatory network in light signaling and identify the novel key regulators in light response.
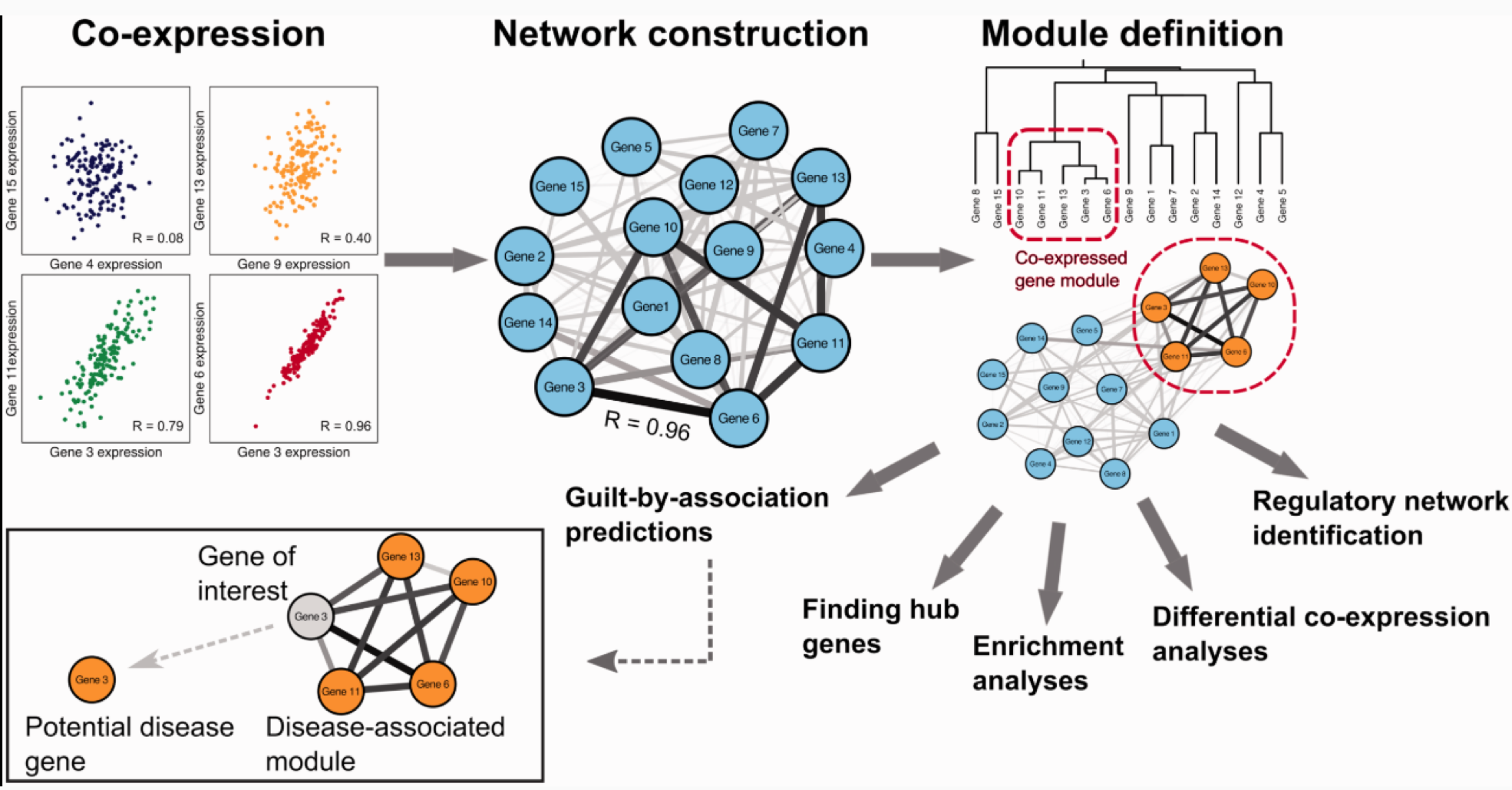
The figure credited to Dam et al., 2017. Gene co-expression analysis for functional classification and gene–disease predictions. Briefings in Bioinformatics
• Project 3: Apply machine learning based gene regulatory networks analysis methods (mlDNA/random forest) using R and Python combined with large sequencing datasets in Arabidopsis to identify the light signaling novel related genes based on expression characteristics patterns.
Future research
I would like to focus on functional genomics analysis and apply this towards researching to improve human being (including human diseases, crops productivity and agricultures)
My ultimate goal is to build a solid knowledge and experience in computational skills as they relate to understanding genetic variants, especially the effects of genomic variants on gene function and signaling networks.
Ultimately, I seek to develop innovative methods with functional genomics data.
